(iCeMS Carlton Lab)
Georgia Kafer
Georgia Kafer
The post doctoral researcher Georgia Kafer and her team found out that a specific chemical modification that happens in DNA helps to repair DNA damage. This finding is hoped to lead to a new way of cancer prevention in the future. When she told us about the research, she said that it was "the most enjoyable" of all the research projects she has worked on.

Please share with us the significance of your manuscript in terms of its research achievements, impact, or uniqueness.
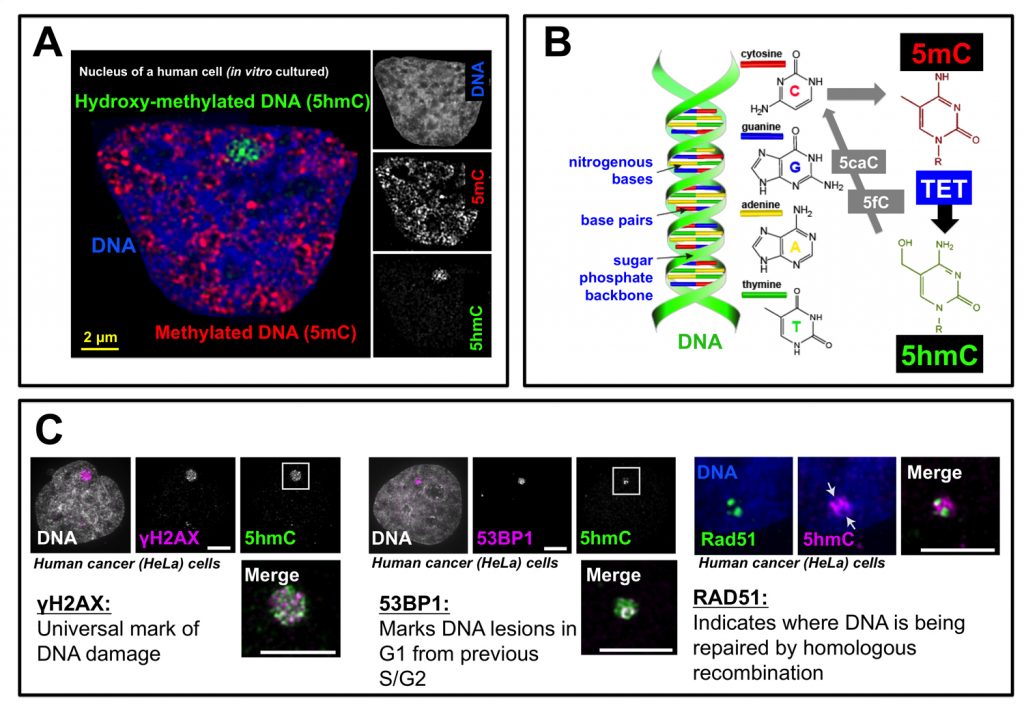
Our genetic information is encoded in DNA. DNA can be modified with small “tags” known as methylation (mC). In 2011 researchers found that there are different types of mC tags, one of which is hydroxymethylcytosine (hmC). For the last 5 years, researchers have been trying to figure out why DNA gets tagged with hmC. Some work has shown that hmC helps genes to be expressed and others have identified that a loss of hmC is associated with cancer. Our research showed for the first time that the hmC tag is associated with damaged DNA. We were able to use very high-resolution microscopy to show that hmC exists at damaged DNA sites in a variety of human and mouse cells. This discovery has far reaching implications for both cancer research and for research investigating how cells repair damaged DNA.
What would you say is the most exciting or “I made it!” moment during this research project?
As with most research we made this discovery almost by “accident”. We were originally trying to develop a method to live-image hmC dynamics. To be confident in our live-imaging results we needed to know what the “normal” pattern of hmC was in the cell nucleus. When I looked at hmC using antibody-based methods in HeLa cells (a very common cancer cell line) I was very surprised to find that there were large “spots” of hmC. These hmC spots had never before been reported in the literature – but they reminded us very much of the sort of spots that some cells (particularly cancer cells) have which are associated with large regions of damaged DNA. We quickly ordered new antibodies that we could use to label damaged DNA and did dual staining to see if our hmC spots localized with the damage spots. They did. Perfectly. It was definitely a very exciting moment and I may have done a little “happy dance” in the dark microscope room before running to find my principle investigator!
Please tell us about the biggest challenge or problem you experienced while conducting your research. How did you overcome it?
Time was, like always, one of the biggest challenges. Research relating to hmC is very competitive as it is a relatively new “hot” area of chromatin biology. DNA damage was a research topic I only knew about superficially, so I had to spend a lot of time familiarizing myself with this new field. I had to learn on the fly – which included making some mistakes – to perform experiments common to DNA damage researchers but quite foreign to me. One particularly memorable challenge was creating a “home-made” system to induce laser-line damage to cell nuclei to test to see if hmC appeared at “new” damage sites. My principle investigator (Peter Carlton) and I spent many hours working in “production line” conditions trying to get a reproducible system to do this. We succeeded in the end and some of our beautiful “damage lines” made it into the manuscript.
Would you consider this work to be a turning point in your overall research direction? If so, how has your research direction changed as a result of this work?
I have always been quite fluid in my approach to research. I like to follow up on whatever interests me at any given time, so I have worked on topics ranging from glucose metabolism to in vitro embryo culture, mouse placental development, human stem cell differentiation and of course most recently DNA damage. However I do have to admit that I have been quite taken with DNA damage research. The DNA damage response of a cell is quite beautiful in how organized it is; yet fascinating in how complex the response may be via the incorporation of many redundant and compensatory mechanisms. Overall I definitely feel that my experience over the last 2 years has persuaded me to try and pursue DNA damage as a research focus. If I can mash this new research love with my old love of embryology and stem cell research then I will be a very happy scientist.
Please describe the current situation of your career. What is your current position? How has the knowledge and experience gained at iCeMS influencing your career?
I completed my PhD almost 5 years ago in Australia, and I am currently a postdoctoral research associate in the Carlton Lab here at iCeMS. Our research is reasonably well funded (though more funding is always welcome!) and so I foresee myself working on this research topic for another year at least. Working at iCeMS has made my “research life” quite easy thanks to the wealth of resources available to me and access to programs designed to “accelerate research” – which was instrumental to the quick turnaround of our discovery into a publication. At iCeMS I have made several professional connections that I truly believe will last throughout my research career and which I will be able to draw upon in future projects. While I am very much enjoying my time in Japan, unless I get a very good offer here I think that one day I will return to work in my native country of Australia. I am sure that if this day comes then my affiliation with a well-regarded research-nexus such as iCeMS will assist me in my future career aspirations.
*All the information on this page, including the researcher’s affiliation, is current at the time of the interview.
Paper information
5-Hydroxymethylcytosine Marks Sites of DNA Damage and Promotes Genome Stability
Georgia Rose Kafer, Xuan Li, Takuro Horii, Isao Suetake, Shoji Tajima, Izuho Hatada, Peter Mark Carlton
Cell Reports
Published: February 2015
DOI: 10.1016/j.celrep.2016.01.035