A New Approach to Gene Therapy: Targeting Mitochondrial DNA
JSPS Postdoctoral Fellow, Visiting Researcher at RIKEN, Visiting Researcher iCeMS (Taniguchi Lab)
PhD student (Sugiyama Lab, Namasivayam Lab), at the time working on this research
Takuya Hidaka
Takuya Hidaka
iCeMS Sugiyama Lab and Namasivayam Lab are conducting chemical-biological research using pyrrole-imidazole polyamides (PIPs). These compounds can bind to targeted DNA sequences to regulate the expression of disease related genes or control the replication of mutated DNA. During his Doctoral studies, Takuya Hidaka developed a new compound that can reduce mutated mitochondrial DNA in live cells.

Please share with us the significance of your manuscript in terms of its research achievements, impact, or uniqueness.
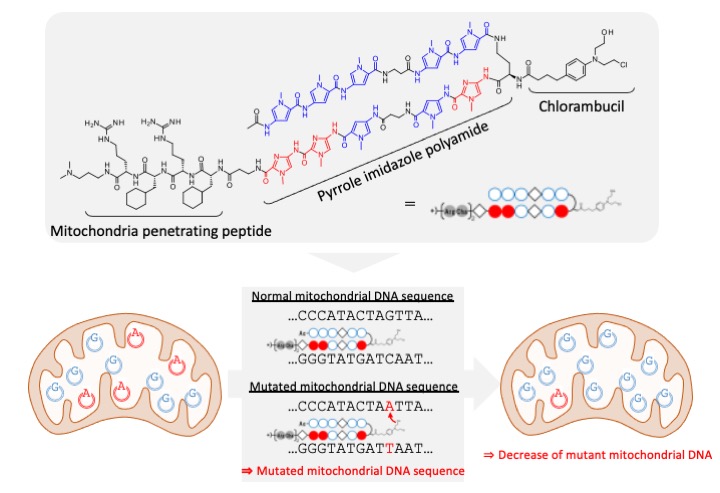
Our cells contain organelles called mitochondria which produce the energy needed for cellular activity. Mitochondria have their own DNA (mtDNA) distinct from the nuclear DNA. Mutations in the mtDNA can cause mitochondrial diseases marked by abnormal mitochondrial function. To cure mitochondrial disease, the mutated mtDNA should be removed from the cell. Until now, genetic engineering using enzymes to cut strands of the mtDNA has been applied to achieve this, but the introduction of foreign DNA such as viral vectors into the cell is required.
In this study, we have developed a molecule that can selectively alkylate mutant bases in mitochondrial DNA by combining a pyrrole-imidazole polyamide with a mitochondrial-permeable peptide and a DNA alkylating agent (chlorambucil). By treating cells with this molecule, we succeeded in reducing the percentage of mutant mitochondrial DNA and proposed a new approach to gene therapy using chemical compounds.
Please tell us what was the most gratifying or inspiring moment for you during this research project.
In order to investigate the amount of mutant mitochondrial DNA after compound treatment, we had been analyzing mitochondrial DNA extracted from cells through quantitative PCR. However, the data varied greatly no matter how many times we tried. We were having trouble obtaining data that could be statistically analyzed and we realized that the alkylated mitochondrial DNA had been fragmented by heating during the PCR. When we analyzed the data after recovering from the alkylation, the decrease in mutant mitochondrial DNA, which had been buried due to the variability of the data, began to appear in a statistically significant form. Making one small realization and devising a plan we were able to overcome a hurdle that had been troubling us for months. I was very happy when I got the final piece of data for my paper.
Please tell us about the biggest challenge or problem you experienced while conducting your research. How did you overcome it?
Developing a compound that shows the targeted effect in living cells was a big challenge in itself. Because of the complex structure of cells, we must consider many possibilities. For instance, has the compound entered the cell or mitochondria, has it reacted with DNA or proteins other than the target, or has the compound degraded in the cell. We prioritized the factors which seemed to be particularly influential, and in the end, we found that the reactivity of the alkylating agent and the position of the mutant base relative to the compound were important. However, this took a long time, more than three years since the 2017 report on which this study is based (J. Am. Chem. Soc 2017, 139, 8444-8447). In fact, due to my degree application, I had come to the point where I thought, “If the next experiment doesn't work out, I am giving up on this research.” Fortunately, I was able to write a paper on another sub-theme, so I was able to complete this research without being bound by it. I think that working on the main theme for a long span of time and the sub-topic for a short span of time in parallel was very helpful in maintaining my motivation for the main theme.
Would you consider this work to be a turning point in your overall research direction? If so, how has your research direction changed as a result of this work?
There was a long period of no progress in this research, and I often wondered if I should continue with this theme. Whenever I stopped to think about whether to continue or not, as long as the possibility was not zero, I persevered taking small steps forward. I believe that this experience and the academic knowledge I gained in the process will help me grow as a researcher and will be a great support for my future research life.
Please describe the current situation of your career. What is your current position? How has the knowledge and experience gained at iCeMS influencing your career?
I received my degree in March 2021, and now I am a JSPS Postdoctoral Fellow at RIKEN and a visiting researcher for iCeMS Taniguchi Lab, which has allowed me to continue to study at iCeMS. I am developing new methods for molecular and cellular biological analysis using microscopy and microfluidic technologies, which is a complete change from the chemical biology research I did while I was a student. I feel that it is only in the interdisciplinary research environment at iCeMS that I can experience such diverse fields of research within the same institute. As I am just beginning to shape my career, I would like to take advantage of this environment to foster my own science that is not limited by research fields and carry it with me to my future career.
*All the information on this page, including the researcher’s affiliation, is current at the time of the interview.
Paper information
Targeted Elimination of Mutated Mitochondrial DNA by a Multi-functional Conjugate Capable of Sequence-specific Adenine Alkylation
Takuya Hidaka, Kaori Hashiya, Toshikazu Bando, Ganesh N. Pandian, and Hiroshi Sugiyama
Cell Chemical Biology
Published: August 2021
DOI: 10.1016/j.chembiol.2021.08.003