Decoding and Recoding the Epigenome
At iCeMS, the fundamental and theoretical, biology and chemistry, technology and research are intricately intertwined. We had a chance to sit down with two scientists trying to understand the genome through different lenses.

Ganesh Pandian Namasivayam
Junior Associate Professor/PI
Born in Tamil Nadu, India. Graduated with a PhD in applied bioscience from Niigata University. Since 2010, he was a member of the Sugiyama Lab at iCeMS and began his own Group in 2018.
Yuichi Taniguchi
Professor/PI
Born in Gifu, Japan. Graduated with a PhD from Graduate School of Engineering Science, Osaka University. He was a postdoctoral fellow at Harvard University until 2011, and a Unit Leader at RIKEN before joining iCeMS.
Communication Design Unit (hereafter known as "CD Unit") From the perspective of epigenetics, or your area of specialty may be slightly different, can you tell us about your research background?
Taniguchi My background is biophysics, a little different from epigenetics. Physics is not only to derive some equations but also to produce some concepts to effectively characterize very complex mechanisms. Towards these concepts, our lab is developing techniques based on many fields’ techniques including imaging microscopes, genetic analyses, computational simulations, bioinformatics, and machine learning. We are searching for the best technology to investigate the mechanisms.

Pandian My background is in biotechnology. I started to do my PhD in the Life and Food Sciences department and there I worked with the immune system of kaiko (silkworm in Japanese). I studied why silkworms don’t get HIV or other microbial diseases. During my study, I reared silkworm a lot and I noticed that they largely varied in size, and it was not suitable for my research because they should be similarly sized. I did not know why they became like that and noticed that food plays a crucial role. It motivated me to start my research around this topic and led to my interest in epigenetics. Before my PhD I didn’t even know that there is a phenomenon called epigenetics as my background was different. There I understood that, okay, genetic code alone is not enough to cover gene expression or the blueprint of the kaiko. Maybe something above it is governing it and I wanted to learn more about epigenetics. This was the starting point for me to come here to iCeMS and start as a postdoc.
CD Unit Can you explain your own research a bit to Taniguchi-san?
Pandian Sure. I'm very interested in genome modality about how the genes are expressed, which has been the topic of my curiosity from my PhD time. This is about knowing the epigenetics and the codes that decide which gene is on and off. My research is to have a closer look and create biomimetic epigenetic codes to control therapeutically important and cell fate controlling genes. In other words, saying in a very simple way, we are all made from letters (A, T, G and C) in DNA.
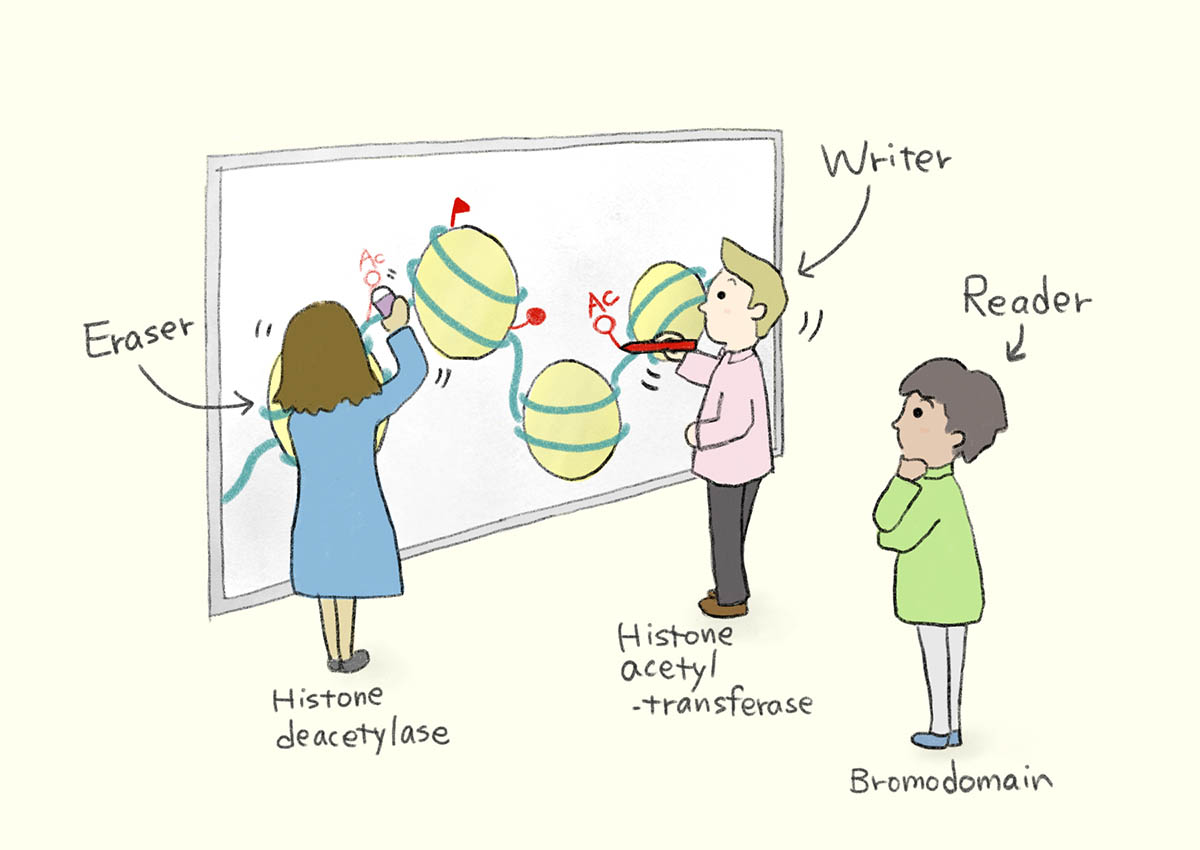
The letters are edited by the epigenetic enzymes that can be broadly classified into three types based on their functions, 1) reader like the bromodomain protein family and 2) eraser like the histone deacetylase (HDAC) enzyme, and 3) writer like the histone acetyltransferase (HAT) enzyme.
Some drugs are known to go and interact with these editors and artificially alter the epigenetic programs that decide which letters are written (switch ON) and unwritten (switch OFF). I was a postdoc in Sugiyama lab which specializes in a fascinating molecule called pyrrole-imidazole polyamide (PIP) that can be pre-programmed to read particular sequence of letters in DNA. What we did is, we started conjugating the epigenetic editors to the DNA readers so that the editor knows where to go and edits only in the region of interest. In this way, we could switch ON the stem cell genes, retinal tissue, and even germ-cell genes even in the terminally differentiated cells like skin cells where they are usually in OFF state.
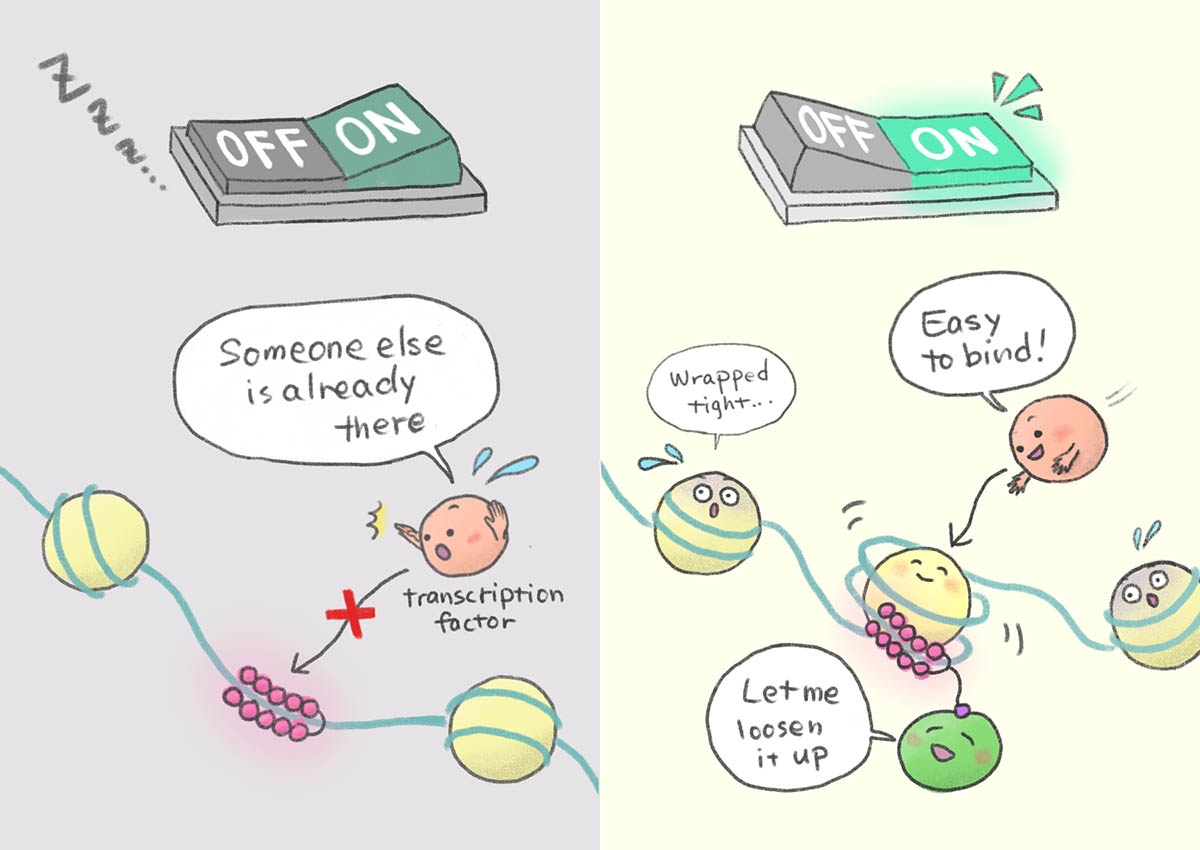
Basically, I intend to switch ON the good genes and switch OFF the bad genes for therapeutic purposes.

Taniguchi Yes, I think it's a very interesting project. For epigenetics I think many molecular biologists are studying that, but they are just using standard techniques and it's just for understanding. But your clinical biology technique can actually modify the status of the cell and it can be directly linked with some medicine. It's a unique approach for your group. I think it's very interesting.
Pandian Thank you. But we still have a long way to go, because playing around with epigenetics is a double-edged sword. So, we have to perfect the technology as much as possible before entering the clinical stage. So, this is still a work-in-progress. To fast-track our journey, we hope to have your support and collaboration. Could you tell me about your exciting research related to epigenetics?
Taniguchi For me, research in my post-doc period is related with epigenetics at the single-cell level. In the research, we measured gene impressions in single cells at the single molecule level, to understand variability and its stochasticity of gene expression states for each cell. By using such a single molecule technique, we could precisely measure the stochasticity in a time-lapse. We found for smaller copy numbers of proteins, stochasticity of gene expression reactions dominates the single cell variability, but for highly expressed proteins, the epigenetic cell state heterogeneity is dominant. It's not like your research, but it's very basic research for understanding single cell epigenetics.
Pandian But this basic research is fundamentally very important. Usually, we picture DNA or epigenetics in just one dimension, and we say, okay, this molecule goes and binds here for this epigenetic event to happen. While we say so, we don’t directly see it but infer it indirectly based on the results. But inside live cells, the epigenetic molecules are in 3D or maybe even 4D. So, I think your technique is essential to have a closer look and we need to know what can happen if we use our or any molecules that can act as genetic switches. Just by the gene expression, we know, okay something is happening. Still, what actually happens or... more insights can be realized from your technique because your information can guide us or others to make better genetic switching tools. So, it's key information that you're giving in real-time dimensions. By the way, this is a question I am always interested in asking you, I saw your very nice cover picture of your article in the Cell. Can you tell me more about that?
Taniguchi Yes. The purpose of the Cell paper is to understand a molecular aspect of the epigenetics. We developed a technique to measure the genome structure at the nucleosome level, which is the smallest structural unit of the genome, constituted by wrapping DNA around the histone molecule.
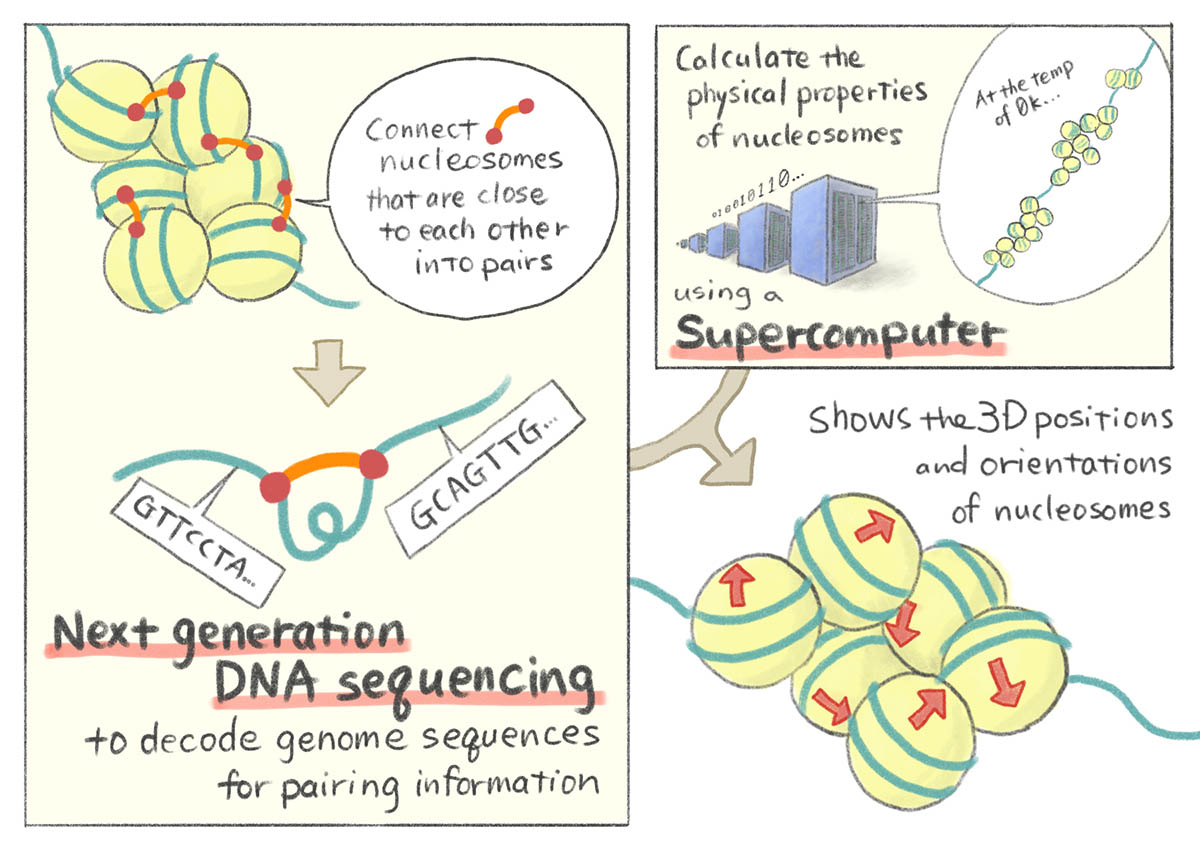
Our resolution is the highest and only our technique can measure the 3D positioning of every nucleosome in a genome-wide scale.
Using that we are trying to understand how genome structure effects gene expressions and histone modification. We're trying to understand the relationship between the genome’s physical structure, chemical reactions, and gene expression in 3D.
Pandian Wow, In real-time, in 3D environment, it's amazing.
Taniguchi Yes. At first, we made such a tool and maybe next time, we will do more research using that technology.
Pandian I'm amazed by your technique again that you're actually seeing, in real time, in 3-dimensions, that's remarkable.
Taniguchi Yes, thank you.
Pandian I'm now interested to know how our biomimetic epigenetic codes behave in 3-dimensions. For example, what is the route? Or how and where our molecule actually goes and approaches in a 3D space. It would be very nice to have a closer look at this process through your technique.
Taniguchi Yes, maybe our 3-dimensional structure, that information, may be good for your design for the DNA probe. We found that DNA structures are various and unique for every genomic locus. Some genomic loci have this structure, and some genomic loci have another structure, so maybe you can make some chemicals to specifically work on some genomic locus.
Pandian That’s right.
Taniguchi It may be, I believe.
Pandian In principle, we can program our molecules to go and attach the letters in DNA like LEGO pieces. But it cannot always happen and we don’t know why? We don’t know whether it's our experimental problem or our design problem. It is challenging to pinpoint one particular cause. Maybe if we see how the whole phenomenon in the natural cellular environment looks like, we may know why it didn't work so that we can know how to make it work?! If we know how our destination i.e., the epigenetic environment really looks like, we can make advanced tools to target them better!
Taniguchi I see. Yes.
Pandian How did you develop the Hi-CO analysis?
Taniguchi How? It's based on a next-generation sequencing experiment and a molecular dynamics simulation technique. That combination is very important. In the experiment, first, we chemically cross-linked the genome and after that, we ligated proximal DNA fragments to each other. Then we sequenced those ligated products with next generation sequencing to find which two genomic sequences are proximal. This produces a huge data set and using that we can find which two genomic loci are close together or far apart. Based on that information we do molecular dynamics simulation to find the best positions, or most reasonable positions, for every nucleosome considering the nucleosome’s physical properties. That's the principle of our technique.
CD Unit Ganesh, can you explain what kind of specific technologies make your research possible?
Pandian Those that can help me to have a closer look at gene regulation inside living cells. While genetic code is deciphered, epigenetic code is not. So far, we have studied the codes around the histone proteins coordinated by the epigenetic enzymes functioning as the readers, writers, and erasers. Previously, I was making molecular tools for DNA. Currently, I'm also interested in RNA epigenetics, like I want to know how the modifications on the letters in RNA can change the way the letters are read or unread so that I can make new tools to modify them on demand.
So far, our PIP technology was used as a proof of concept study to say, okay, this can gene can be switched ON and OFF but not on a translational scale. But recently, we have shown that we can construct biomimetic epigenetic code to synergize cancer immunotherapy by switching ON a gene that can produce mitochondria. Basically, in the fight between the immune cells and the cancer cells, cancer cells are known to grow by sucking the mitochondria from the immune cells like T cells. So, we increased the mitochondrial biogenesis in T cells by targeting the responsible genes at the molecular level so that they can kill the cancer cell better. It will be nice to go more than that.
CD Unit Okay, so yes, can you think of someone who is inside iCeMS doing work related to epigenetics?
Pandian Yes, Dan Ohtan Wang’s lab works on the basic science of RNA epigenetics studying synaptic plasticity. In fact, her work inspired my new ongoing research theme to develop applicable tools for RNA epigenetics. When Ohtan’s former lab member joined my lab, we discussed and thought of a chemical biology approach to decode RNA epigenetics at single nucleotide resolution using a pocket-sized nanopore sequencing device. Nanopore sequencing technology enables the real-time analysis of the conventional letters in DNA (A, T, G and C) or RNA (A, U, G, C) molecules by monitoring the changes in the current pattern using the in-house machine learning algorithms. But there are also unconventional modifications, for example, A can become I (Inosine) or m6A (N6-methyladenosine) which may serve as disease biomarkers, and we usually need sequencing technologies involving large-scale devices to assess them. However, a simple device like the nanopore sequencers is preferrable in a clinical setting if we want to read the modified letters and identify diseases. Using nucleic acid chemistry, we design molecules that can selectively read these letters, we create a traceable change over these therapeutically important letters in RNA molecules, and accurately detect their right place in a transcriptome scale. Basically, we first decode the RNA epigenetics using designer chemical tools and aim to recode them later.
CD Unit I see. Nice, so working at iCeMS gives you the RNA epigenetics ideas. Perhaps there are not many people working on epigenetics at iCeMS? But there are a lot of other researchers out there. Then do you find some potential for collaboration within iCeMS with other researchers?
Taniguchi I think researchers studying epigenetics are not so many in iCeMS or in general, compared with the ones studying gene expressions. But the epigenetics is an upstream of gene expression processes, so when considering the regulation mechanisms, people can get interested in going to epigenetics.
CD Unit Taniguchi-sensei, your research involves various types of analysis, have you thought about the potential for collaborations with other chemistry researchers, not only with biology or epigenetics researchers?
Taniguchi I'm always thinking about it since joining iCeMS. I think it would be interesting to make probes, labeling reagents, etc., since chemistry at iCeMS is so strong, and I need to find a good solution by filling the gap between what I can do and what would be interesting to make. Recently Hidaka-san has come from Ganesh-san’s lab, and he has both (a biology and chemistry) background. I've been talking with him about various possibilities; however, things are always not so easy, but every day we are enjoying the discussion.
CD Unit Daniel Packwood-san and Ohtan-san, I've heard they’ve been doing simulations, for example, modeling the behavior of RNA. Theoretically, do you feel like this (collaboration beyond research fields) is something you could work on?
Taniguchi Theoretically, yes. First of all, we think about what is biologically significant, and then consider the solution. For example, we have considered a probe that would light up when two (molecules) were attached to each other, which would improve the analysis.
CD Unit Imaging-wise?
Taniguchi Imaging-wise. We talked about the possibility, but it turned out to be difficult. There are a lot of things that would be nice to be able to do. We're exploring them one by one, discussing the possibilities.

Pandian Talking about imaging, Sugiyama-sensei has DNA origami technology where he folds DNA in a nanoscale to form a frame within which one can visualize single molecule interactions using atomic force microscopy. I am interested in using them for the real-time visualization of epigenetic events like DNA winding and unwinding over histone proteins and Taniguchi-San may consider using it too.
About AI, while not exactly epigenetics, day and night can alter our body’s biological clock and at the molecular level our gene expression. To map such higher order processes involving big data, I collaborate with Daniel’s lab in iCeMS to develop programming codes. Actually, while we are talking now, Daniel's postdoc and my postdoc who both have completely different backgrounds are sitting together in my lab and developing a machine learning algorithm to decode which genes are ON and OFF in the day and night.
CD Unit So the actual collaboration is happening there. Nice.
Taniguchi Yes, something interesting.
CD Unit Well, yes, you might be able to collaborate in such kind of ways.
Taniguchi Yes.
Pandian I would love to.
Taniguchi Yes, definitely.
Pandian We haven't talked about our collaboration yet, but decoding is a fundamental process in recoding and so Taniguchi-san’s tool will be useful for my research. I can show you what I have and if you think that I have something useful for your research, then that will be great.
Taniguchi Yes. I think it's a good idea.
CD Unit Taniguchi-sensei, you just started your iCeMS life a couple of months ago. Ganesh-sensei, you've being here for a while. You both probably have future vision what you want to do at iCeMS or maybe even the further future, so what do you going to do after this?
Pandian
My future vision is to decode and recode the genetic switches for treating non-communicable diseases associated with epigenetics. Lifestyle changes like diet can affect the epigenetic codes and is known to have significant impact on living things. I’ll tell you my bee story as an example. All bees start their lives as babies in the form of a larva. As per the mysterious rule in the bee world, some of them will have continue to have their diet with royal jelly and becomes queen bee and those who do not have it become worker bee. Likewise for humans, unhealthy lifestyle habits can cause diseases and may also be inherited.
Particularly, post-corona lockdown, lifestyle diseases may increase exponentially as people are not sleeping properly, not eating on time, and eating unhealthy foods. But there is good news. Unlike the irreversible DNA mutation, the epigenetic inheritance can be reversed. I hope to advance my tools to decode and recode the epigenetic marks associated with diseases. I don’t know how much, but at least I want to flatten this peak as much as possible and contribute to a healthy society for my children or grandchildren. To realize this dream and translate our findings, I am also involved in a startup company founded by Sugiyama-sensei.
Taniguchi I see. I think our vision is also similar. At our lab we are trying to develop new approach by integrating many technologies and once it's established maybe we will find other targets and make new concept technology. We have a direction to understand the genome activity in silico based on the structure analysis. If succeeded, we may be able to simulate the genomic reactions and to predict the gene expression regulation. It's very related with Ganesh-san’s research. Really, my future vision is related with his.
Pandian My dream is dependent on his finding. We complement each other. Whether the DNA/RNA in the microscopic scale controls the cells or whether cells at the macroscopic scale control the DNA/RNA has always been like the chicken-egg story. The nucleosome level which Taniguchi-san is interpreting, like decoding, is where the actual control happens, and the decision about the fate of a cell, whether it is going to be a normal cell, or a diseased cell, or a skin cell, or retinal cell – all those decisions take place in that particular place that Taniguchi-san is decoding. What we try to do is to mimic and recode this using programmable molecular codes. Epigenetics, which operates in the mesoscopic scale, also matches well with the founding convictions of iCeMS. So, please decode. Let me recode.
Taniguchi Yes.
CD Unit Yes. That's a nice talk. Thank you so much.
Taniguchi Thank you.
*1 Dan Ohtan Wang: Former iCeMS PI (Until March 2021). Currently the team leader of the Laboratory for Neuroepitranscriptomics at RIKEN Center for Biosystems Dynamics Research.