Shedding Light on Cell Death Signals
Program-Specific Assistant Professor (Suzuki Lab)
Masahiro Maruoka
Masahiro Maruoka
As new cells are born, old cells that are no longer being used meet their end, and thus maintain homeostasis in the living organisms. The lipid called phosphatidylserine acts as a marker when displayed on an old cell's surface and is recognized by phagocytes which then remove the dead cell. In this study, Dr Maruoka, Prof Suzuki and their colleagues shed light on the regulatory mechanism of lipid dynamics which occurs when cells die.

Please share with us the significance of your manuscript in terms of its research achievements, impact, or uniqueness.
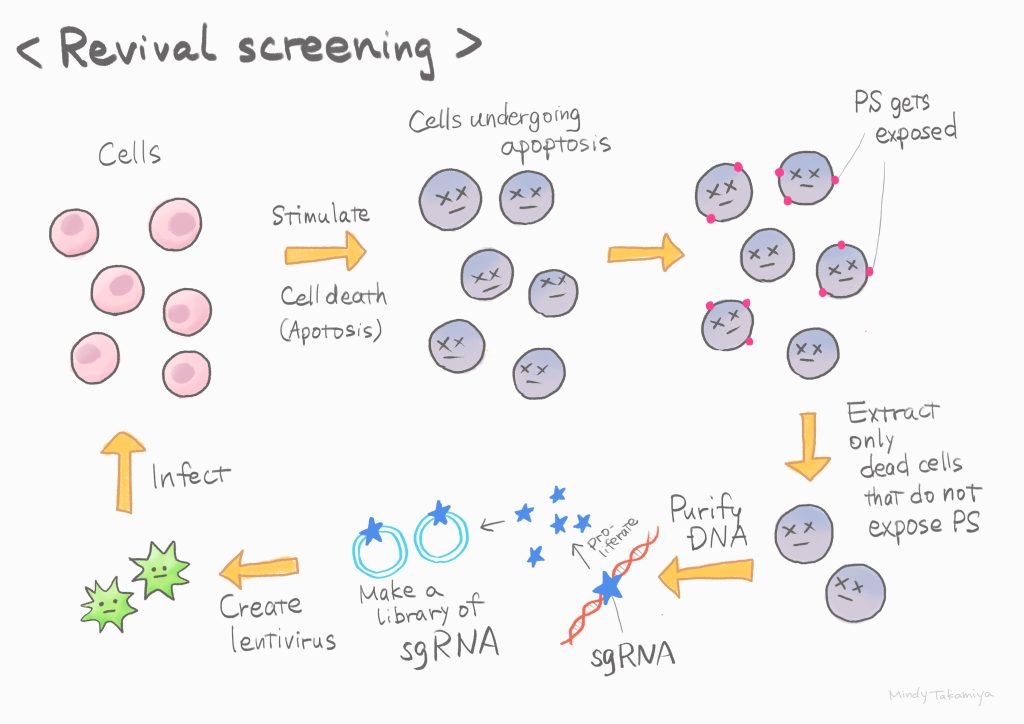
In living organisms, old and unwanted cells undergo cell death (apoptosis) and are removed. During this process, a phospholipid called phosphatidylserine (PS) is exposed on the outer surface of the cell membrane, and phagocytes recognize it and eat the dead cell. In our laboratory, we have identified scramblases that are involved in the exposure of these lipids and have analyzed their functions. In this study, I attempted to identify the regulatory molecule to understand the activation mechanismof Xkr4, a scramblase expressed specifically in neurons. Scrambling can be induced by apoptotic stimulation, but when scrambling activity is used as an indicator in screening, the cells die and are lost. Therefore, I developed a new "revival screening method" using a CRISPR/Cas9 sgRNA library, which enables comprehensive gene disruption, to identify candidate genes even from dying cells. As a result, I identified XRCC4, one of the nuclear DNA repair complexes, as an activator of Xkr4. This result overturns the conventional wisdom that signals for removal of unwanted cells are transmitted directly from the nucleus to the cell membrane. We hope that our understanding of this mechanism will provide new clues to prevent neurodegenerative diseases.
Please tell us about the biggest challenge or problem you experienced while conducting your research. How did you overcome it?
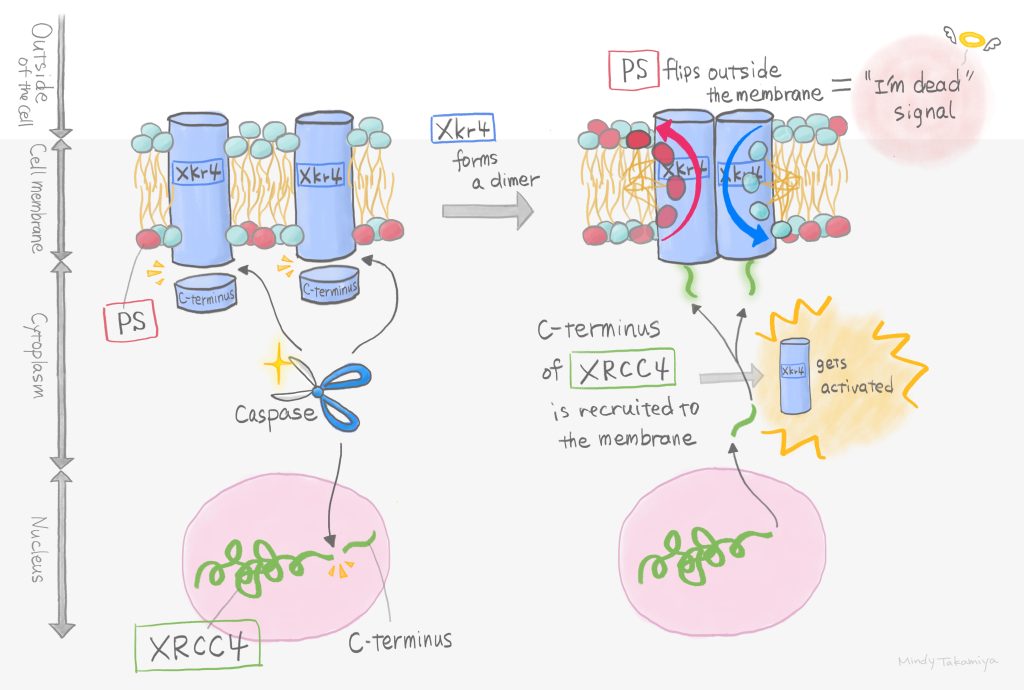
In this study, it was a challenge to set up a new screening system, but I think it was also a challenge to analyze XRCC4, which was identified as a candidate gene. This is because XRCC4 is one of the DNA repair complexes, so it naturally exists in the nucleus. On the other hand, Xkr4 is only found in the cell membrane. Since the localization of these molecules was completely different, following traditional thinking, it was hard to believe that they would work together. Despite our doubts, our analysis revealed that 1) XRCC4 is cleaved by caspases that are activated during apoptosis, 2) some of the cleaved fragments are released into the cytoplasm, and 3) the released fragments bind to Xkr4 on the plasma membrane. I believe that we were able to find the answers to these questions by constructing a logical screening system and carefully analyzing the results one by one without being bound by preconceptions or common sense.
Please tell us what was the most gratifying or inspiring moment for you during this research project.
This was when we proved our hypothesis through direct experimentation. Because a fragment of XRCC4 bound to Xkr4, we hypothesized that this interaction would contribute to the scrambling activity of Xkr4. To prove this, we found the minimal peptide region of XRCC4 required for activation, and when we directly injected the peptide into the cells, we found that Xkr4 was activated without inducing apoptosis. This result proved our hypothesis, and I remember feeling both happy and relieved that our screening results were correct.
Would you consider this work to be a turning point in your overall research direction? If so, how has your research direction changed as a result of this work?
It is believed there are about 20,000 human genes. Until I arrived here, I had assumed that the majority functions of most of the genes were pretty well understood. However, after screening, I not only realized that the majority of functions of genes are not yet understood, in my research, I discovered that even XRCC4, the gene whose function was thought to be well understood, has an unknown function. I think this experience was a very significant turning point in that it made me realize the importance of screening in understanding life phenomena. I will always keep this attitude of trying to investigate life’s phenomena through screening in mind in my future research.
Please describe the current situation of your career. What is your current position? How has the knowledge and experience gained at iCeMS influenced your career?
I continue to work at the iCeMS Suzuki Lab as a program-specific assistant professor. I conduct my own experiments and supervise students. Based on the revival screening we are conducting collaborative research to identify new factors involved in phenomena of interest to researchers in Japan and abroad. At iCeMS, I am fortunate to have the opportunity to collaborate with people from a variety of backgrounds, including materials science, chemistry, biology, and mathematics. In addition, not only do we have access to the latest equipment, but we also have a well-developed support system in the Analysis Center. I am grateful to be able to carry out my research in such a favorable environment, and I will do my best to connect it to my future career.
*All the information on this page, including the researcher’s affiliation, is current at the time of the interview.
Paper information
Caspase cleavage releases a nuclear protein fragment that stimulates phospholipid scrambling at the plasma membrane
Masahiro Maruoka, Panpan Zhang, Hiromi Mori, Eiichi Imanishi, Daniel Packwood, Hiroshi Harada, Hidetaka Kosako, Jun Suzuki
Molecular Cell
Published: March 2020
DOI: 10.1016/j.molcel.2021.02.025